How to use the Arbor webapp for comparative analyses
So, you want to use the Arbor web app to do some comparative analyses?
The Arbor web app is a flexible workflow engine that will let you carry out a wide range of visualizations and analyses. You can upload data files from your computer, or use data from a range of web services.
For this example, I will walk you through the steps you need to use the Arbor web app. I will set up a basic analysis of a correlation between two continuous characters using independent contrasts.
Get started
You will need the anolis example data files, anolis.phy and anolis.csv, from [Arbor datasets] (/arborCollections/docs_datasets/). Save those files somewhere on your computer.
Now open the Arbor web app. Your screen should look something like this:

If you want, you can log in here; we will post more information about Arbor user accounts later. For now, you don’t need to log in, you can do the analyses for this example without an account.
Upload your data
Near the bottom of the Arbor web app screen you will see a blue box that says “Browse or drop files.” This is how you can get your data into Arbor. There are two options: either you can click the box and browse to the file you want, or you can drag the file onto the box and let go. Arbor should recognize what type of file you have added (in this case, tree or datafile) and store them in the correct way.
Using this method, load in both anolis.phy and anolis.csv. If you want to make sure that your upload worked, you can use the dropdown menu that says “Save or download data.” If you have properly loaded the files you will see them appear in that list.

This is what my screen looks like after uploading anolis.csv - note that anolis.phy is also in the “Save or download data” dropdown, as I uploaded it just before.
Note: Since you have not added your files to any collections, they will appear like this:
- anolis.csv
- anolis.phy
You may also see the same files stored in a collection, depending on how your web app is set up. If you see versions of those stored files from the “datasets” collection, for example, they will look like this:
- anolis.csv (datasets)
- anolis.phy (datasets)
Those stored files will work fine for the exercise, but try to use the ones you uploaded so that you can get the hang of using your own data in Arbor. Also, since these datasets are uploaded but not saved, if you refresh your browser or even accidentally go to a different page, the datasets will be gone when you come back. If you want to save your uploaded data then you should login and make a personal collection.
Select the collections you want
For this exercise, you will use functions from two arbor collections, “base” and “phylogeny.” All available collections are listed to the right side of the Data Management tab.
To gain access to functions in those collections, click the little eyeball next to “base” and “phylogeny.” Your screen should now look like this:

Navigate to the “analysis” tab
You are done with the Data Management tab, and will need to move to the “Analysis” tab to carry out your analysis. Click on the Analysis tab to switch. Your screen should now look like this:

Try out the picCorrelation function
We can now do some comparative analyses! Arbor has two types of analyses: functions and workflows. Later, we will build a workflow - but actually, for this problem, there is a single function that we can use to get output immediately.
The function we need is called picCorrelation, and it is part of the phylogeny package.
First, select this function (in the Select analysis dropdown menu). You can inspect the function by clicking “Show script” (this example is in R). You should see this:
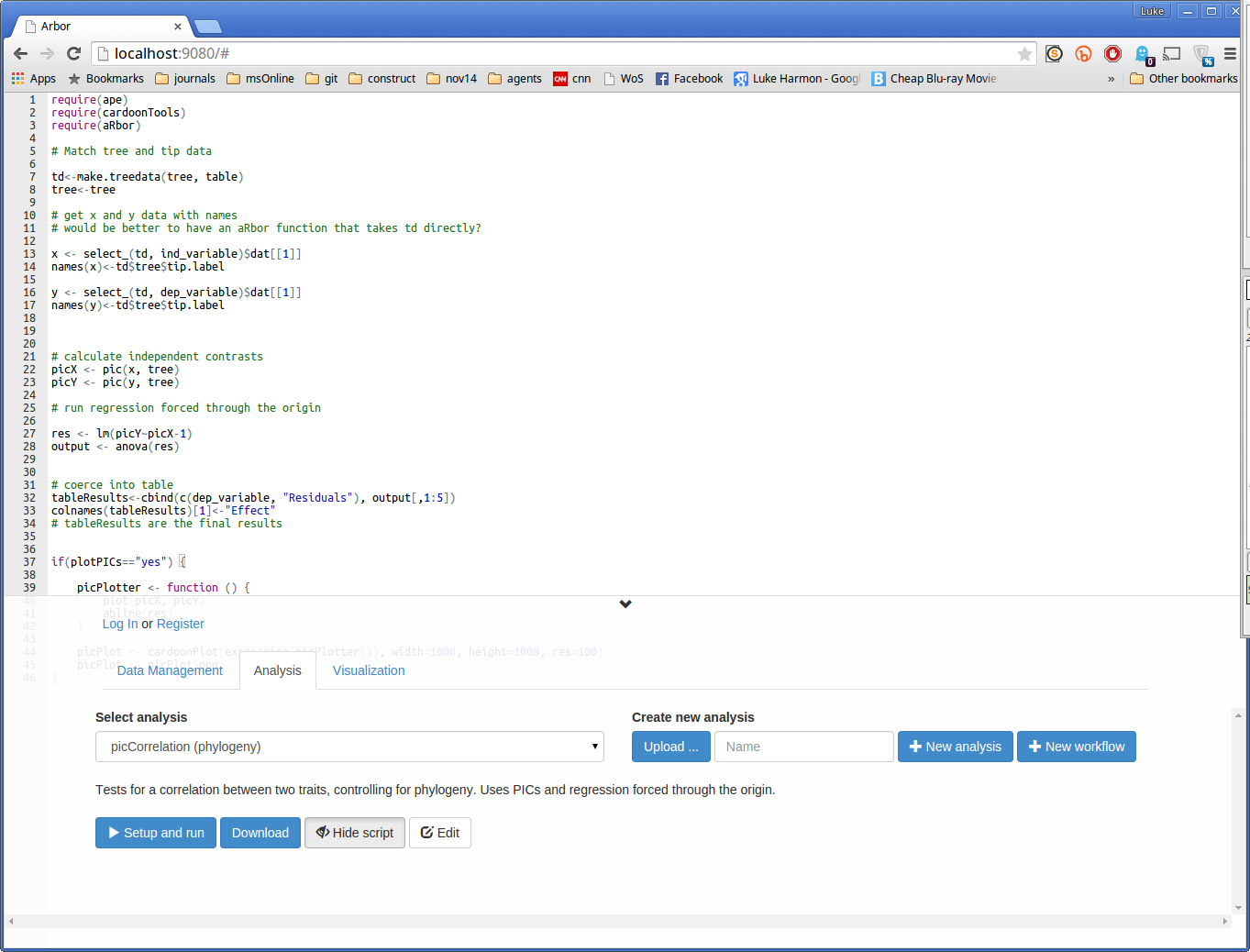
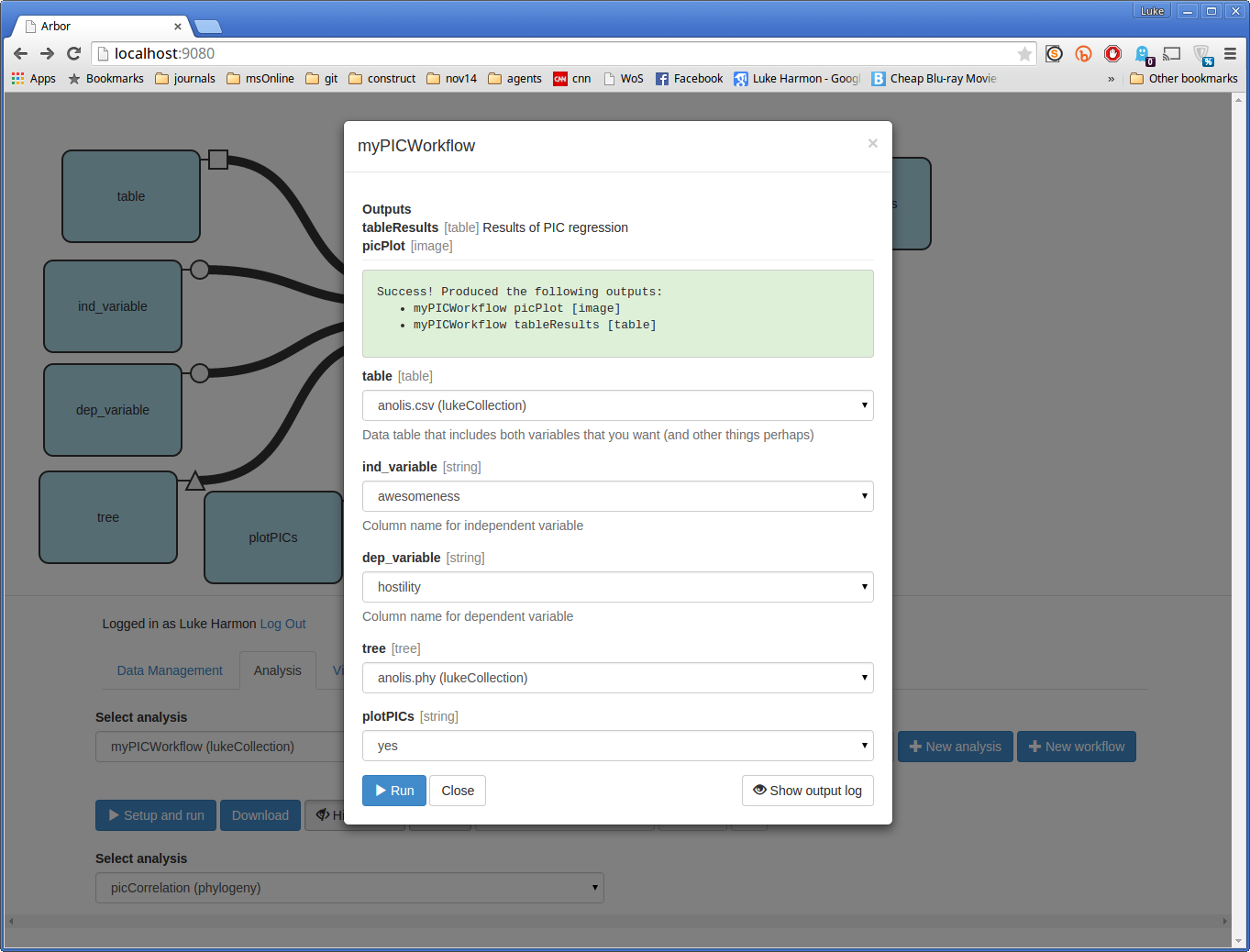
You can now run the function by clicking the small blue button that says “> Setup and run.” This brings up a dialog box. Fill in the missing details in this box to analyze hostility (dep_variable) as a function of awesomeness (ind_variable). Choose the table and tree that you entered earlier, and set “plotPICs” to “yes.” Your form should look like this:
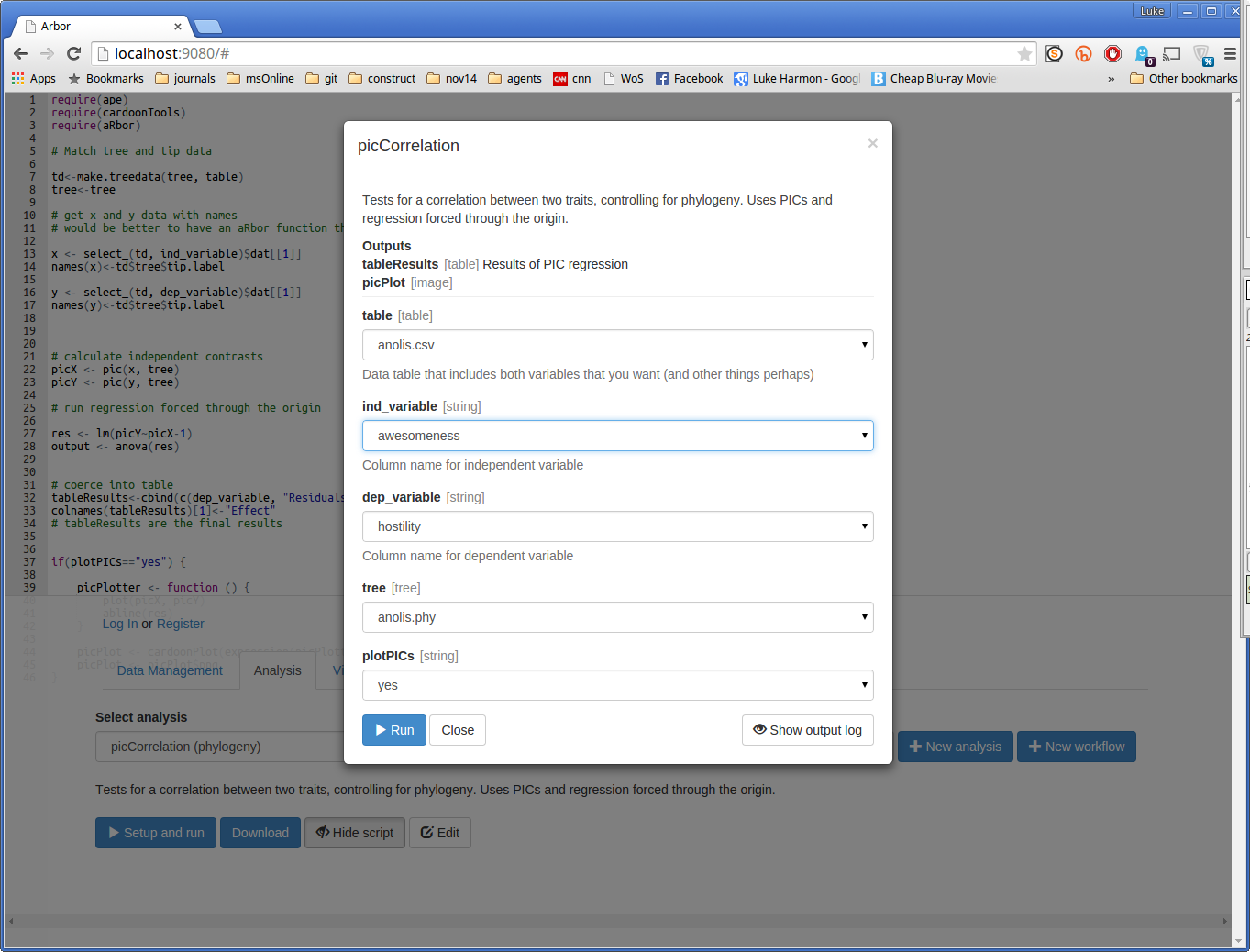
Now press “> Run” and see what happens. If your run works, you should see a success message:
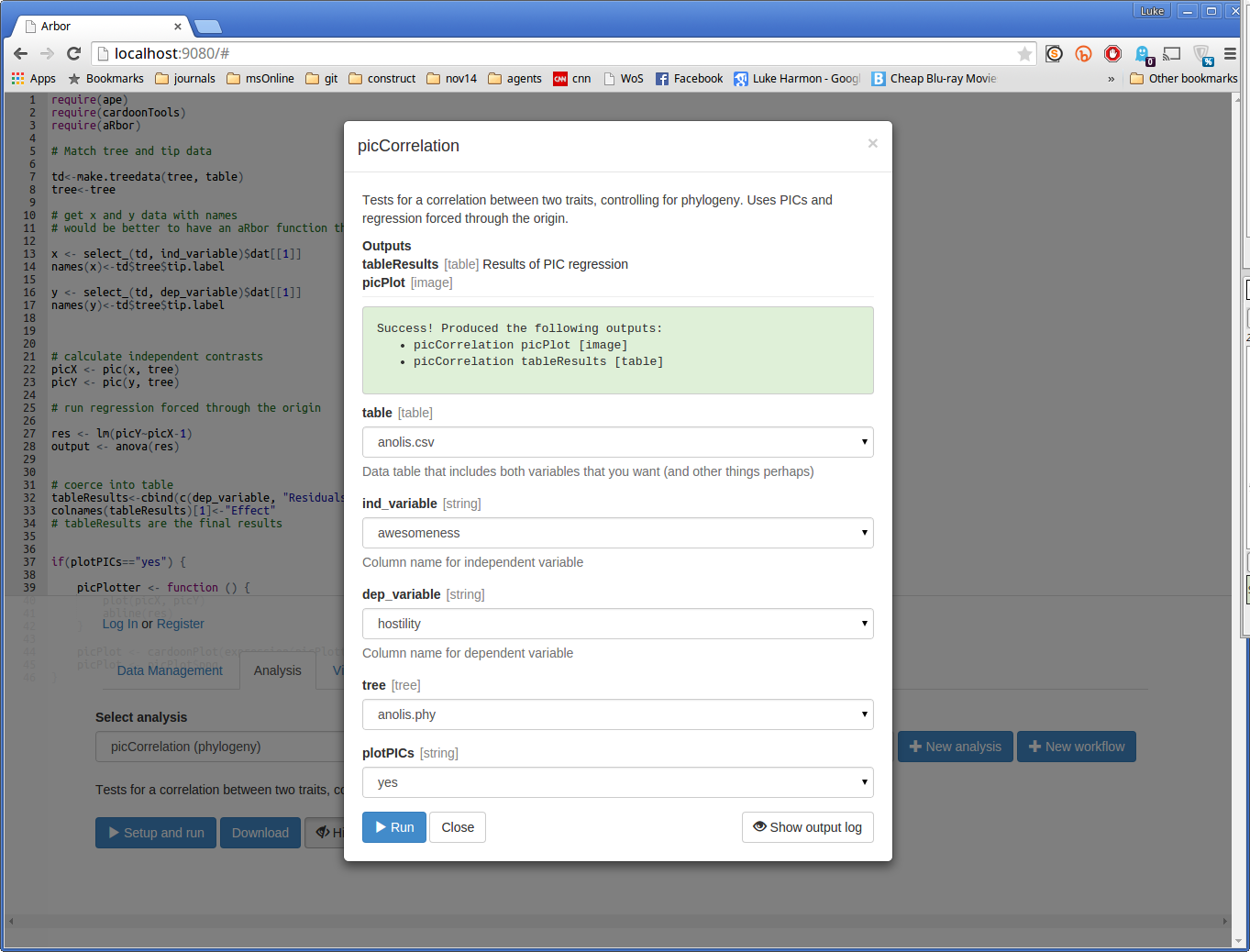
Here, Arbor is telling us that the function had two outputs:
- picCorrelation picPlot, which is an image
- picCorrelation tableResults, which is a table
Let’s have a look at the outputs.
Visualize the outputs of the picCorrelation function
To see the outputs of an Arbor function (or workflow) we can go to the Visualization tab. Click on it now. Your screen should look like this:
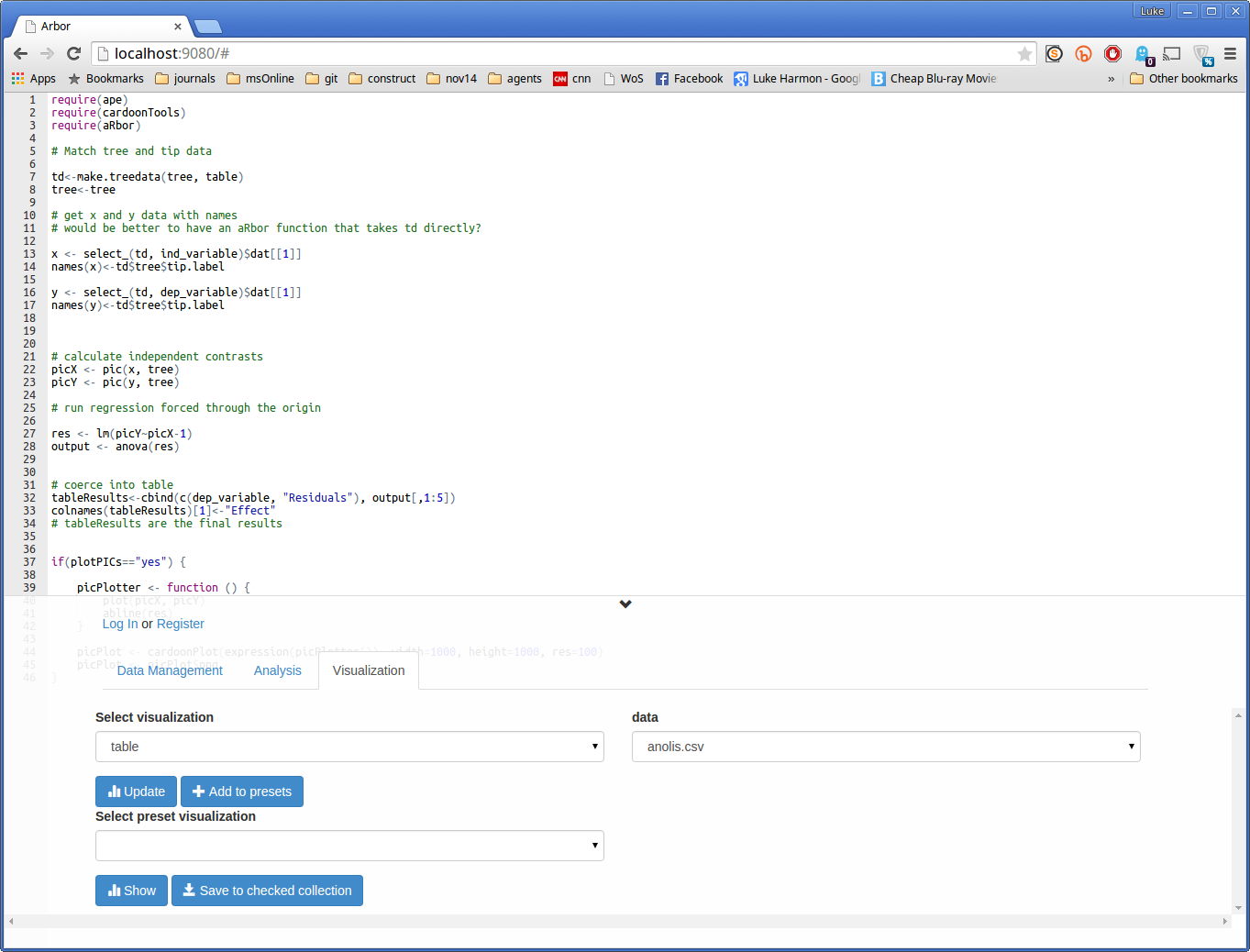
Note that the top part of the screen did not change, but the bottom tabs and buttons did. That’s ok! Keep going.
We can look at the plot first. Set the “Select visualization” dropdown to “image”, and you should see your output, called “picCorrelation picPlot”, appear under “data.” To activate this visualization click on the blue Update button. The result should be a plot.
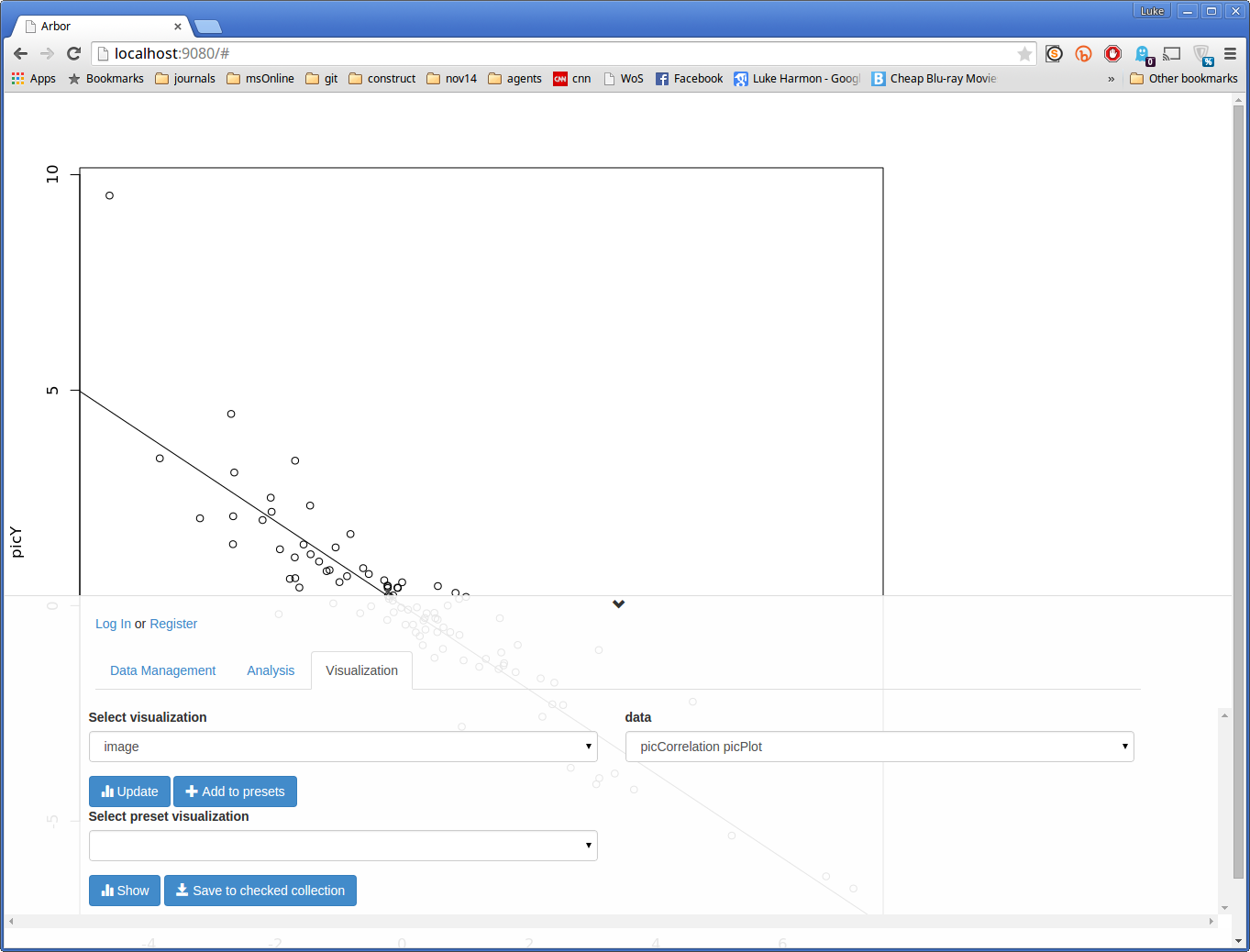
(You can see more of the plot by clicking on the small downward-facing arrow above the “Log In or Register” section of the screen).
Next, let’s see some numbers. Change “Select visualization” to “table”, and “data” to “picCorrelation tableResults.” Hit update again to see an ANOVA table summarizing the results of the phylogenetic regression.
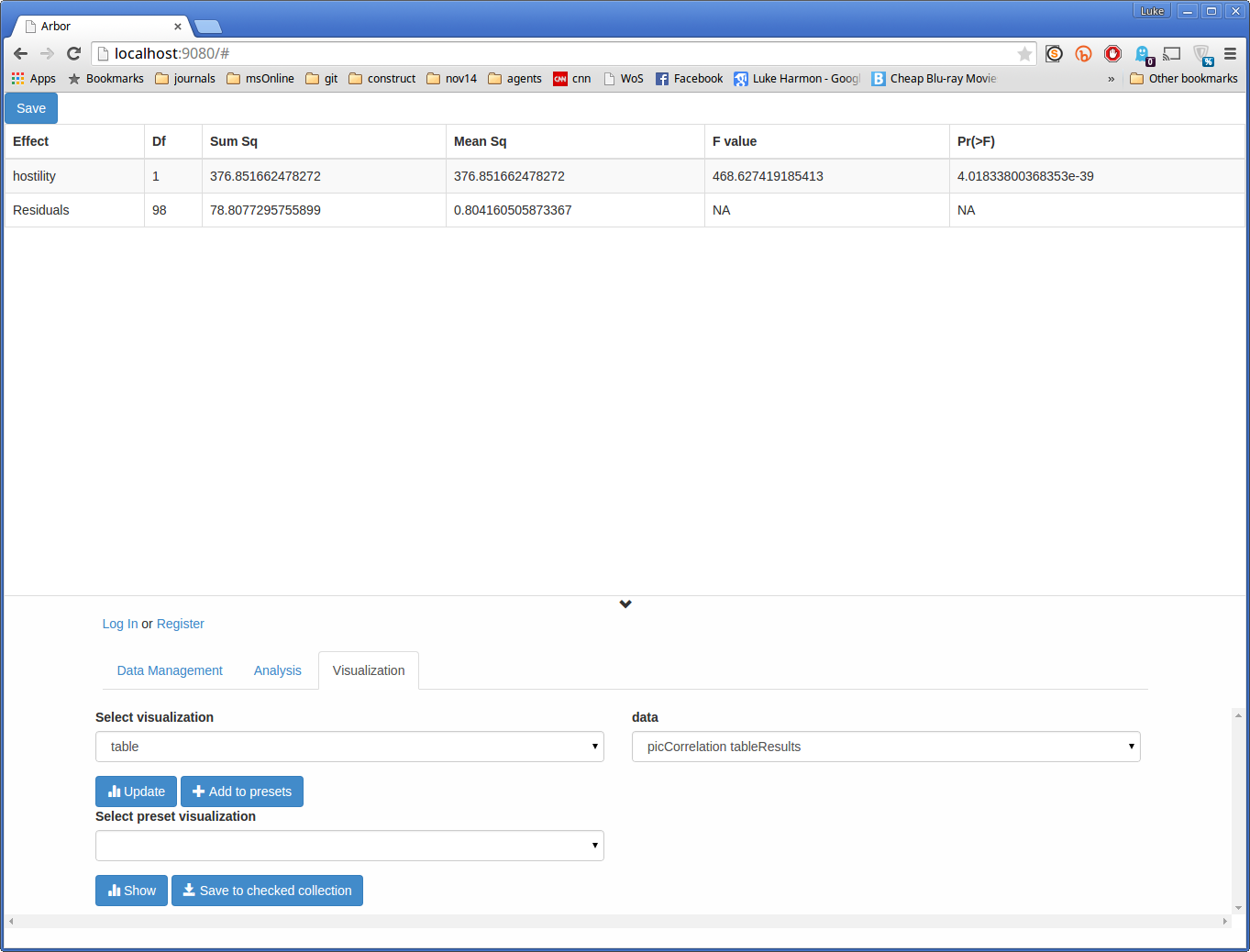
The p-value is the top number in the column denoted “Pr(>F).”
Build a pic workflow
Now let’s try to harness the real power of Arbor by building a simple visual workflow for our analysis. For this part to work you need to be logged in! If you are not logged in you can run existing functions and workflows but you cannot create new ones. You can create an account in Arbor quite easily.
Follow the instructions on this page to create an account and start a private collection. Make sure to click on the “write” tab in your new collection, then come back here and finish the tutorial!
OK I will assume you did that. Now, go back to the “Analysis” tab. To start your workflow, type a name into the box under “Create new analysis,” then click the “+ New workflow” button. I am going to call my analysis “myPICWorkflow.”
Once you have done that, your new (empty) workflow will be available under the “Select analysis” tab. To confirm that it is there (and empty!), select it, and click “Show script.” As we’ll be building this workflow, you can also click “Edit.”
Now all of that empty white space is a blank canvass for your new workflow.
We will build our workflow in three steps:
- Select the function that we need, picCorrelation
- Link the function inputs that we need
- Link the function outputs that we need
For step one, use the drop-down menu labeled “Select analysis.” Find the picCorrelation function - it should be labeled “picCorrelation (phylogeny)” to let you know which collection it belongs with. Then click “+ Add to workflow” and you should see this function appear in the workflow as a block.
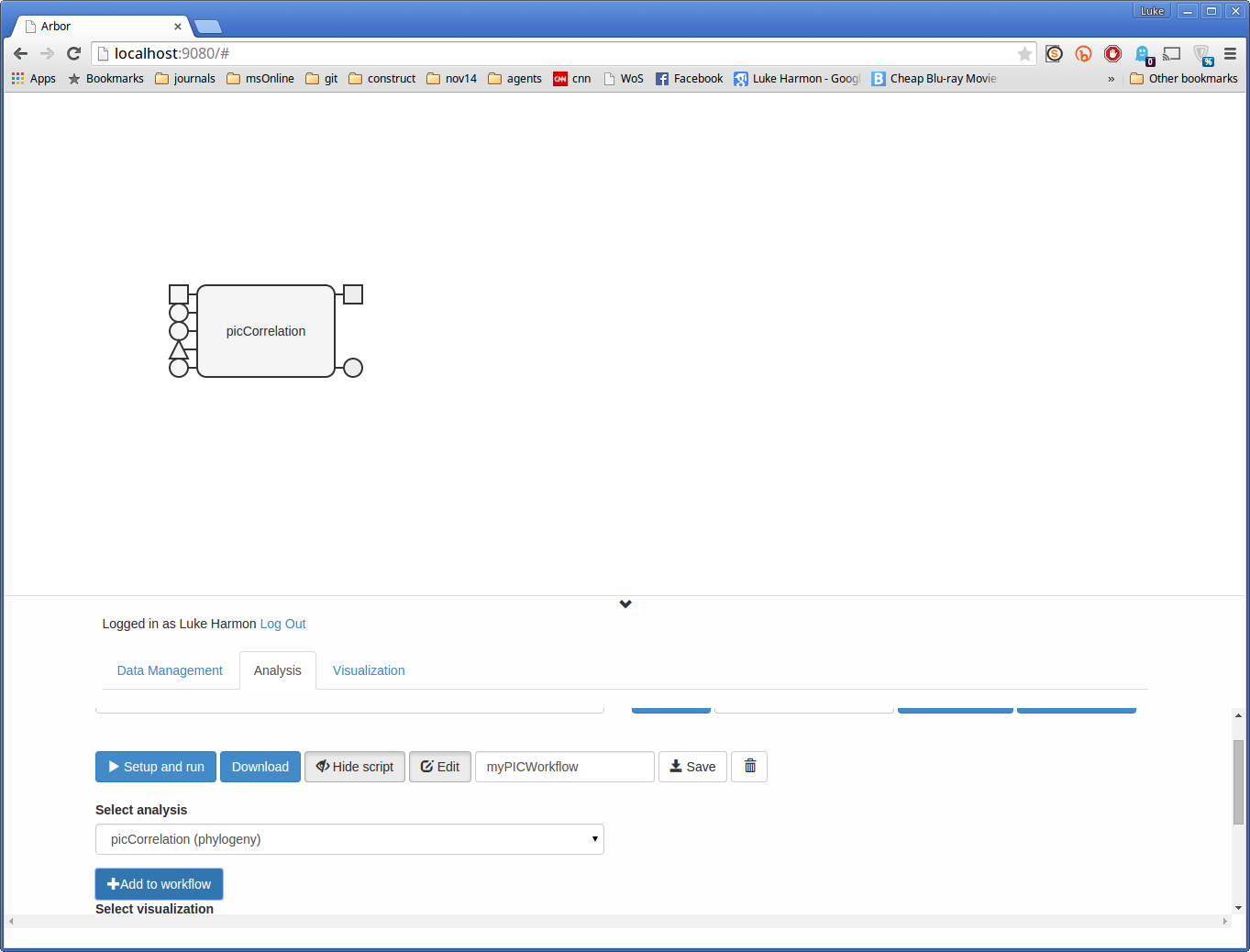
The function itself is a large rectangular block. The things sticking out on the left are inputs - you can see their type by hovering the mouse over each one. Squares are data matrices, triangles are phylogenies, and circles represent text values - typically, these circles control how the function works.
Now let’s make some inputs. Click on each of the input tabs for your function to automatically generate function input boxes.
These input boxes are blue to differentiate them from grey functions. They also only have output tabs, no inputs. All of the input boxes that you create will have to be specified by the user when you run the function, as you will see in a second.
In more complicated workflows later, you will use outputs from previous steps as inputs for the next - but don’t worry about that for now.
Beware! Sometimes new input blocks pop up right on top of function blocks - so it might look like nothing has happened. Don’t panic - just click on their center and drag them out of the way.
You can also rearrange the inputs to look nice.
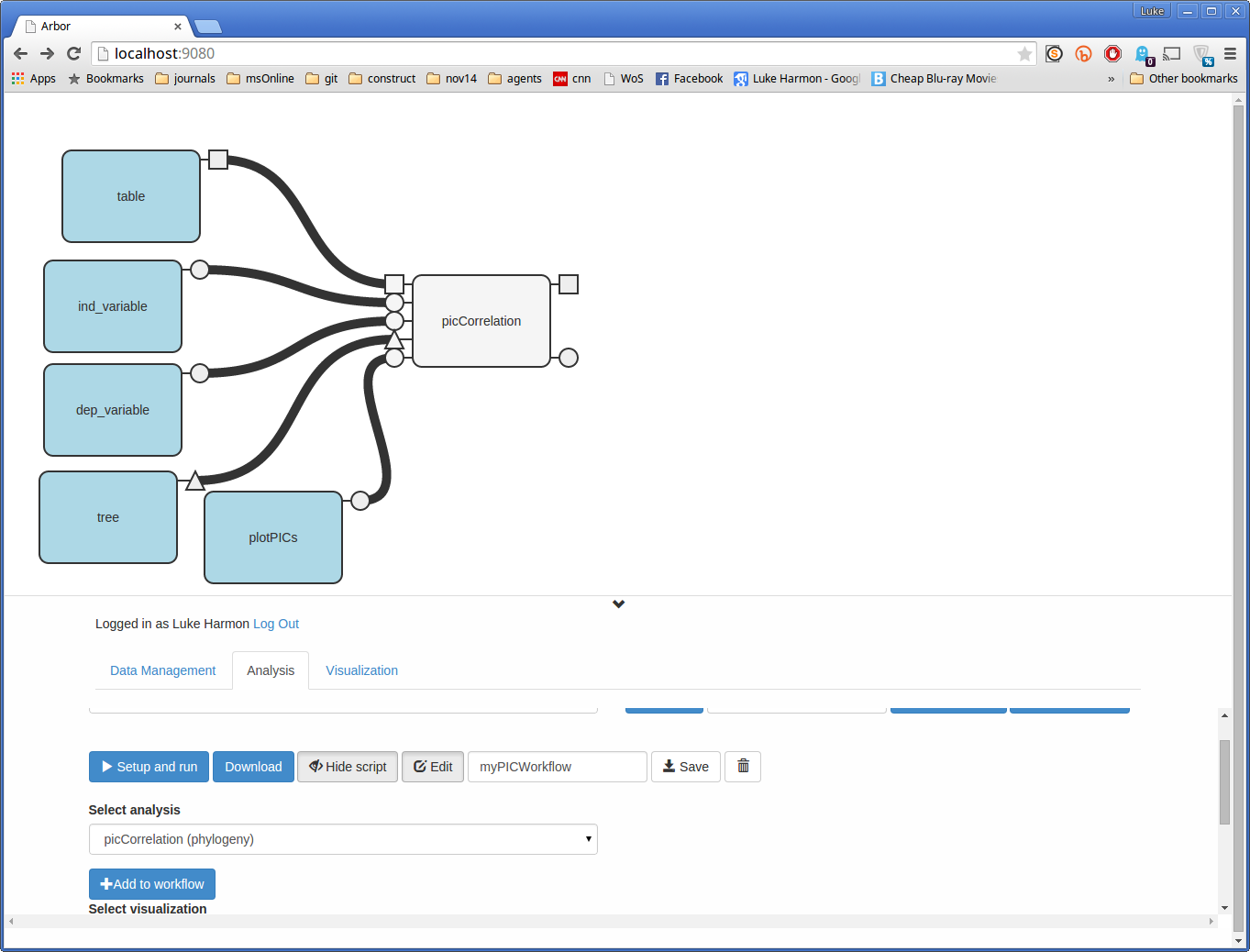
Note: every tab for function input needs to be connected to something! Either a block that is specified by the user, or another part of the workflow.
Almost done - we only need to connect some outputs to see our results. We can do that in the same way that we created inputs - by clicking on function tabs, in this case on the output side, and creating output boxes.
Output boxes are the same color as input boxes but they only have connectors on the left side. Your workflow will create a different output object for each of the output boxes that you create.
You can arrange these to look nice too.
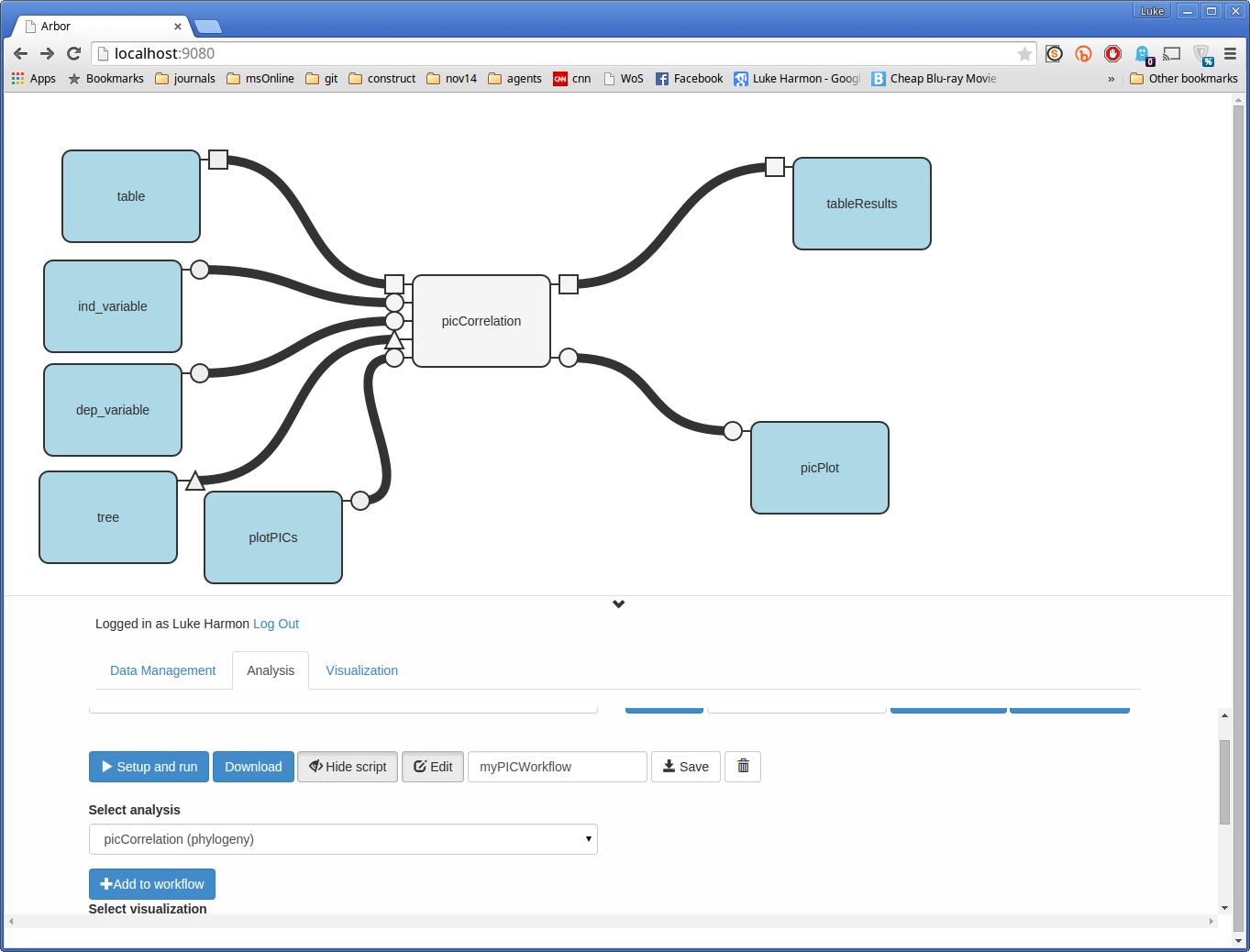
Run myPICWorkflow workflow
Once you have built your workflow, you need to save it before running.
If you just click run then Arbor will run the workflow as it was the last time you saved - which might not match the workflow you see on the screen!
Once you have saved, then click “Setup and run” to start the workflow. You will again have to specify inputs and options. Let’s do the same analysis we did before.
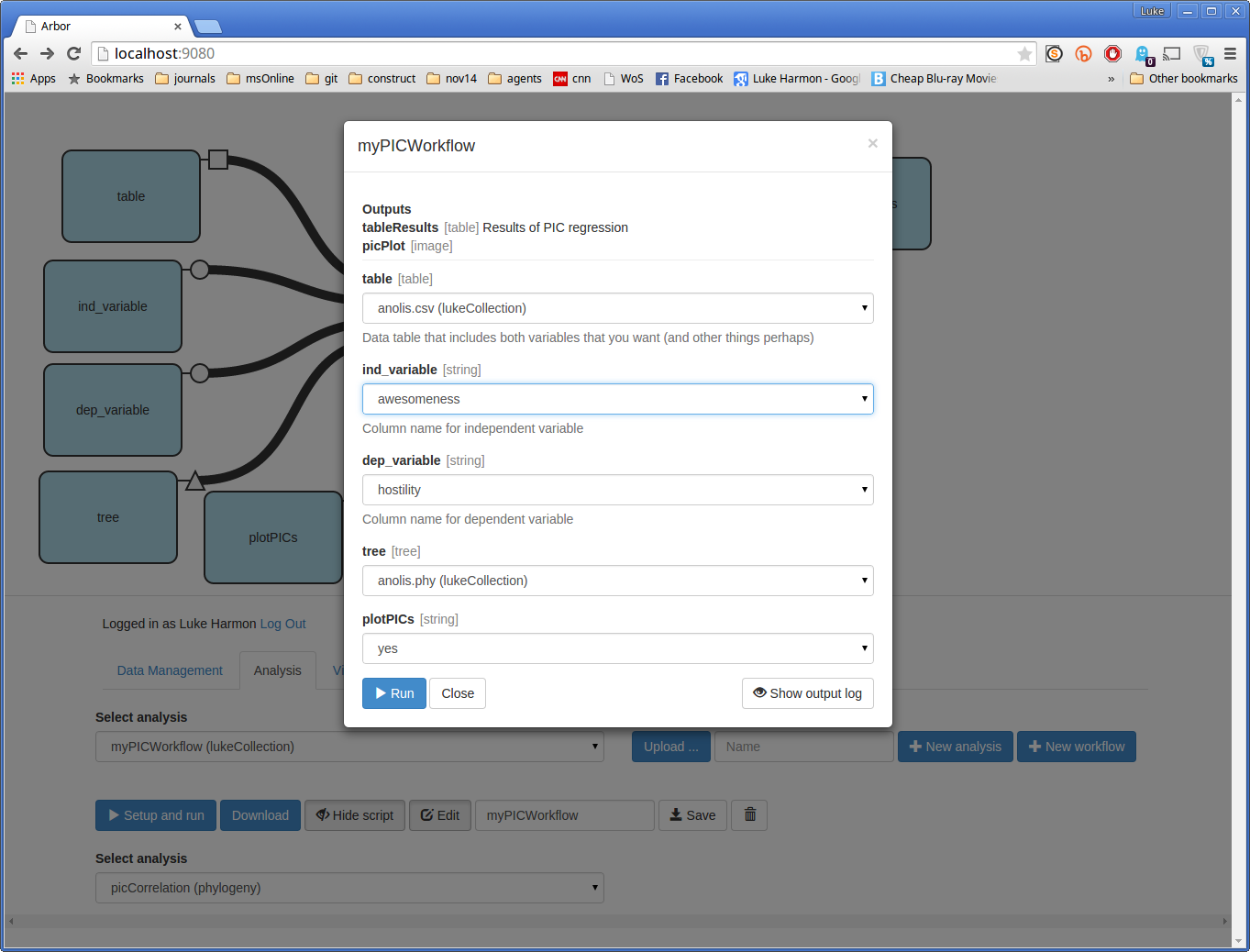
Now click “> Run.” If your workflow works you will see a green success message.
You will see that we have two new output objects, one is an image and one is a table. You can visualize both of them in the same way as above, using the “Visualization” tab.
If you made it this far, nice work - you built and ran an Arbor workflow!